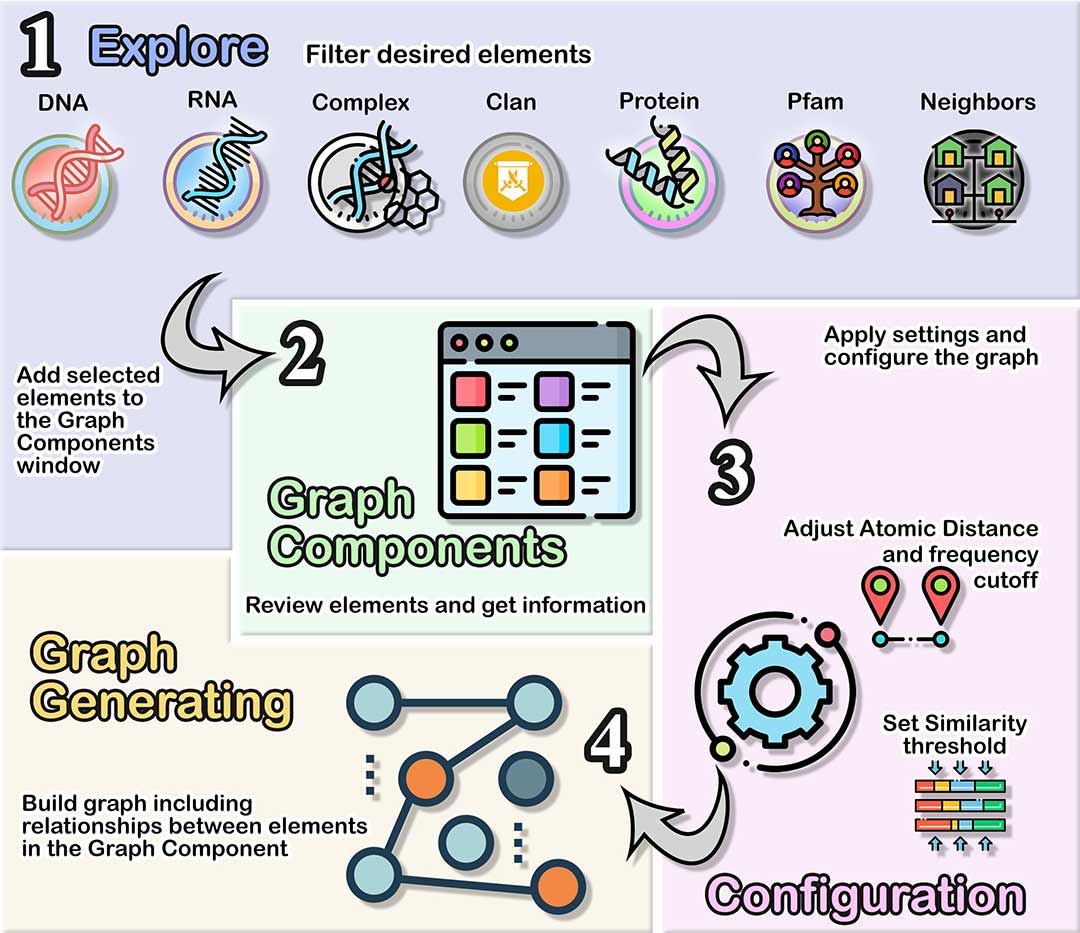
Nucleic acid and Protein interaction network designer
Configuration
Include
| Minimum Length | Maximum Length | |
|
|
||
|
|
||
|
|
||
|
|
||
|
|
Protein interaction with:
Distance Calculation Method
|
|
|
Similarity
({{Explorer.Criteria.Configuration.Sim}} %)
Distance Cutoff
|
Distance (Å)
|
|

|
Explore
Empty boxes consider all items.
| UID | PDB | Sequence | Len | Description | Score | Local % | Global % | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| {{pdb}} +{{rna.PDBs.length-4}} |
{{rna.sequence.substring(0, 10) + '...' +
rna.sequence.substring(rna.sequence.length-10,
rna.sequence.length)}}
|
{{rna.sequence.length}} |
{{rna.auth_desc}}
|
{{rna.Score}} | {{rna.globalSim.toFixed(2)}} | {{rna.localSim.toFixed(2)}} | |||||
Result set is empty.
Empty boxes consider all items.
| UID | PDB | Sequence | Len | Description | Score | Local % | Global % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| {{pdb}} +{{dna.PDBs.length-4}} |
{{dna.sequence.substring(0, 10) + '...' +
dna.sequence.substring(dna.sequence.length-10,
dna.sequence.length)}}
|
{{dna.sequence.length}} |
{{dna.auth_desc}}
|
{{dna.Score}} | {{dna.globalSim.toFixed(2)}} | {{dna.localSim.toFixed(2)}} | |||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||
Result set is empty.
Empty boxes consider all items.
| UID | PDB | Sequence | Len | Description | Score | Local % | Global % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| {{pdb}} +{{protein.PDBs.length-4}} |
{{protein.sequence.substring(0, 10) + '...' +
protein.sequence.substring(protein.sequence.length-10,
protein.sequence.length)}}
|
{{protein.sequence.length}} |
{{protein.auth_desc}}
|
{{protein.Score}} | {{protein.globalSim.toFixed(2)}} | {{protein.localSim.toFixed(2)}} | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Result set is empty.
| UID | Protein | Nucleic Acid | #Complex | Distance(p,na) <= {{ ComplexResult.Tag['Distance']}} Å | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| {{comp.uid_pro}} | {{comp.sequence_type}} | {{comp.uid_na}} | {{comp.frequency}} | {{comp[ComplexResult.Tag['FieldName']].toFixed(0)}} | ||||||||||||||||||||
|
Protein Description:
{{comp.desc_protein}}
Protein PDBs:
{{p}}
{{comp.sequence_type}} Description:
{{comp.desc_na}}
{{comp.sequence_type}} PDBs:
{{p}}
Protein Sequence:
{{comp.seq_protein.length}} amino acids
{{comp.seq_protein}}
{{comp.sequence_type}} Sequence:
{{comp.seq_na.length}} nucleotides
{{comp.seq_na}}
|
|||||||||||||||||||||||
Click buttons to reload desired neighbors of the nodes added in the graph
{{Neighbor.SelectedNode.sequence_type}}: {{Neighbor.SelectedNode.uid}}
Interactions
{{pair.uid}}
Similar macromolecules
{{uid}}
{{sim.GlobalSimilarity.toFixed(1)}}
Network
The graph is empty
RNA:
{{GraphInfo.NodeInfo.RNA}}
DNA:
{{GraphInfo.NodeInfo.DNA}}
Protein:
{{GraphInfo.NodeInfo.Protein}}
Clan:
{{GraphInfo.NodeInfo.Clan}}
Pfam:
{{GraphInfo.NodeInfo.PFAM}}
Similarity:
{{GraphInfo.EdgeInfo.Similarity}}
Nucleic Acid Protein Binding:
{{GraphInfo.EdgeInfo.NAProBinding}}
Protein Pfam:
{{GraphInfo.EdgeInfo.ProteinPfam}}
Clan Pfam:
{{GraphInfo.EdgeInfo.ClanPfam}}
Connected Compoenents:
{{GraphInfo.ConnectedComponents}}
NC: Not converged (limitations are depends on server resources and
nodes` count)
| Node | Degree in subgraph | Closeness Centrality | Betweenness Centrality | Eigenvector Centrality | Degree in Total |
|---|---|---|---|---|---|
| {{gi.Node}} | {{gi.SubgraphDegree}} | {{(gi.ClosenessCentrality*100).toFixed(2)}}% | {{(gi.BetweennessCentrality*100).toFixed(2)}}% |
{{(gi.EigenvectorCentrality*100).toFixed(2)}}%
{{gi.EigenvectorCentrality}}
|
{{gi.TotalDegree}} |
Keywords in the graph
The Graph Components container is empty. Add some nodes first.
| Graph Components |
Preview - Total {{GCs.length}} items.
{{el.uid}}
|
| Export Network | The Graph Components container is empty. Add some nodes first. |
| Dynamic representation | Show The graph must be rebuilt first |
The Graph Components container is empty. Add some nodes first.
| {{cd.uid}} {{cd.type}} | {{cd.title}} | {{cd.description}} | |
{{cd.context}}
|
|||
Graph Components
{{Explorer.Items.length}}
nematzadeh.sajjad@gmail.com